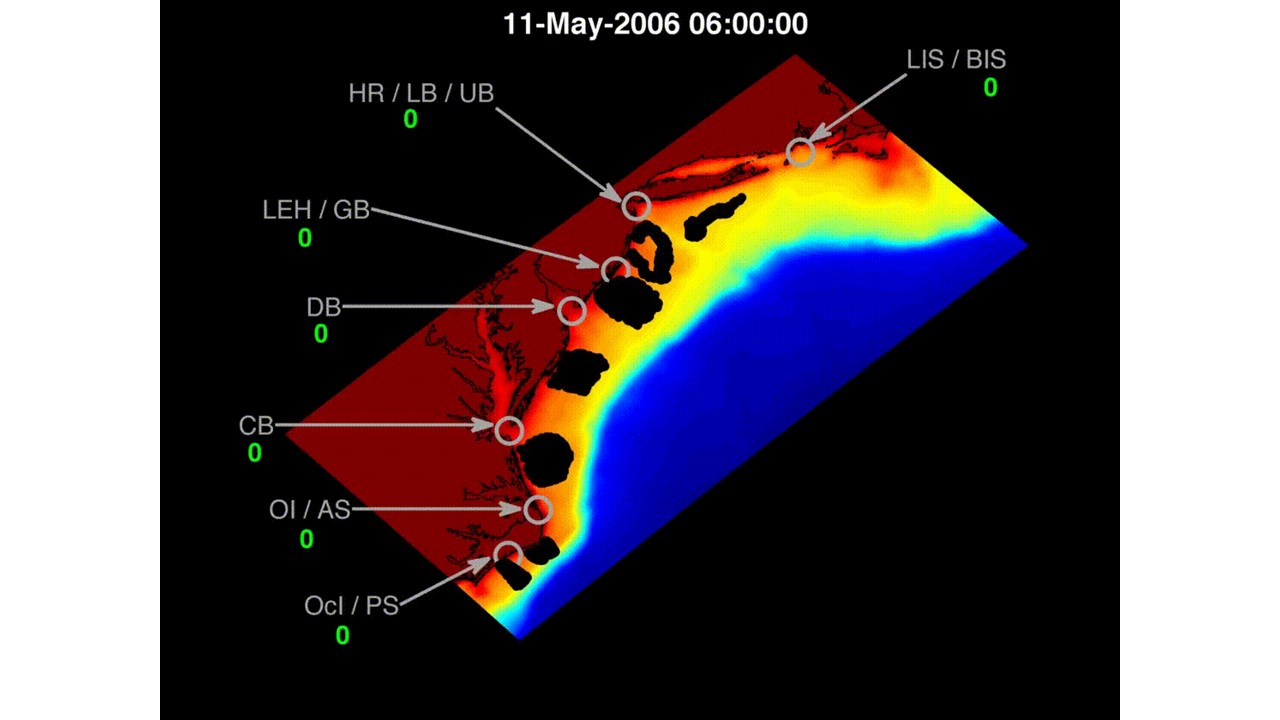
Model results from a physical biological model tracking larval menhaden in the mid-Atlantic Bight (Contribution of Menhaden Nurseries project)
Project
| Contributors | Affiliation | Role |
|---|---|---|
| Miller, Thomas | University of Maryland Center for Environmental Science (UMCES/CBL) | Principal Investigator |
| Kinkade, Danie | Woods Hole Oceanographic Institution (WHOI BCO-DMO) | BCO-DMO Data Manager |
Modeling combined an existing operational ROMS model (ESPRESSO) with an existing larval tracking algorithm (LTRANS). Methods & Sampling description for detailed methods.
The modeling combined an existing operational ROMS model (ESPRESSO - http://www.myroms.org/applications/espresso/) with an existing larval tracking algorithm (LTRANS - http://northweb.hpl.umces.edu/LTRANS.htm). Linking of the hydrodynamics (Espresso) and larval biology coupling was completed offline, i.e. flow solutions were computed and stored prior to being used as sequential input to a custom written MATLAB file which simulated the biology. The MATLAB file conducted simulations which updated growth, and position (using LTRANS) every 60 s, and then abundance and recruitment to potential nursery estuaries were assessed every 2 hours. We used MATLAB for all post-LTRANS analyses. Analysis of these trajectories revealed particle interactions with the coastline, including potential nursery sites, yielding likely recruitment patterns.
The model simulated the fates of 2,100 particles over 90 d runs. Each simulated particle was associated with an internal amount – that is the number of individuals it represented – and thus each particle represented a “super individual.” Initially each particle represented 10,000 individuals, thereby making the initial number tracked to be 210 million larvae. The internal amount for each particle was reduced to reflect mortality every 2 hr during the simulation.
A sample animation of the movement of simulated menhaden larvae is available here.
The internal amount of each particle is represented by its shading – initially the internal amount of 10,000 is represented as black, with the particle becoming whiter as it represents fewer and fewer larvae.
Example simulation runs are available on line at http://hjort.cbl.umces.edu/FishOce.html
Model output consists of X,Y,Z,T and abundance of tracked particles in an individual-based model. Spatial coordinates are defined in meters and time in seconds.
The impact of multiple nursery areas and adult age structure on the population dynamics of marine fishes (Contribution of Menhaden Nurseries)
Description from NSF award abstract:
Many marine populations exhibit complex life histories in which larval and juvenile stages are spatially separated from adults. This is the case for many coastal-spawning, estuarine-dependent fishes which utilize multiple estuaries as nursery grounds to ensure that recruitment failure in any single estuary does not translate to total recruitment failure at the population level. For these species, the location and timing of spawning is believed to regulate the pattern of supply of larvae to potential estuarine nursery areas. Furthermore, many of these species exhibit age-dependent coastal migrations which increase in amplitude with age. Thus, there is the potential that changes in the age structure in the population can affect the pattern of supply of larvae to nursery areas and structure the pattern of recruitment. The investigators will carry out an integrated empirical and simulation approach to study the sources, patterns and consequences of larval supply to estuarine nursery areas for Atlantic menhaden (Brevoortia tyrannus) along the East Coast of the US. The first goal will be to quantify the contribution of these nursery areas to coast wide recruitment. Juvenile menhaden from nursery areas from Massachusetts to Georgia will be sampled and the microchemical constituents of their otoliths will be characterized. These chemical signatures will be used to assign the nursery affinities of adult menhaden in the coastwide population. The investigators will test the null hypothesis that the Chesapeake Bay remains the most important source of recruits to the population. By determining the nursery affinities of adults from different year classes in the population they will assess whether the contribution of nurseries varies or has shifted over time. The second goal is use a population model linked to an individual-based coupled physical-biological model of recruitment to evaluate whether the known age-dependent migrations of adult menhaden are sufficient to cause the observed shifts in the distribution of larval menhaden that seed potential nursery areas. The simulation model will assist in evaluating mechanisms behind observed changes in the distribution of juvenile menhaden.
This work will contribute to the fundamental understanding of the regulation of spatially-structured marine populations. The last decade has seen the range extension of several estuarine-dependent marine species with dispersive larvae and the long-term recruitment decline of others. This integrated research program seeks to explore the effects of population demography, oceanographic circulation, and nursery site diversity on subsequent population dynamics. Given the documented changes in habitat quality in many estuarine nursery areas, and the anticipated impacts of climate change on oceanographic circulation, distributional changes in individual species are likely to become more common. Moreover, given the pivotal role that many estuarine-dependent species play in many marine ecosystems, understanding distributional changes will have direct consequences for the structure and function of the ecosystems to which they belong. The project will also train young scientists in areas of research (quantitative fisheries ecology, physical oceanography) for which there is current a national need.
Note: This project is an NSF Collaborative Research project.
| Funding Source | Award |
|---|---|
| NSF Division of Ocean Sciences (NSF OCE) |
[ table of contents | back to top ]